Information Theory in Molecular Biology
(Updated 12 April 2022)As you know, this website is about a new interpretation of biology (the science of life) in terms of information and its processing. The main theme is that living is information processing, at all levels from molecular up to the whole planetary system of bio-geo-chemical cycles. Well, in the pages introduced in this thematic introduction, we concentrate on the molecular level. That means at the level of DNA, RNA and their relatives and proteins and small molecules that may be used to signal or to help in the running of a cell.
Information molecules
The first and the most developed application of information theory in biology concerns the molecular basis of genetics and, by implication, of reproduction and evolution by natural selection. This is not surprising, since it is in the genetic code that we find the most obvious connection between biology and information. The very word ‘code’ implies an intuitive understanding that a DNA sequence constitutes stored information and its translation, transcription or replication is information transfer, or communication.

DNA and RNA are called information molecules because their purpose in life is to hold information, pure and simple. We can interpret that in terms of their causal relationship with all the other molecules composing a living cell (see our blogpost on that). Remember the principle here is that information is an essential ingredient of causation (see the physical basis of causation to understand that). Following Farnsworth (2022), efficient cause (what makes things happen) is the effect of embodied information constraining physical forces. The embodied information acts as formal cause (the constraints) determining the composition and behaviour of the fabricated molecules inside a cell.
In the case of DNA information is embodied in the nucleotide sequence of course, but it cannot be allowed to directly constrain the molecular forces to produce efficient cause. That is because the information it embodies must be used selectively, with the right pieces of information deployed at the right time, i.e. the right formal causes engaged at the right time, not all of them at once. To achieve that, the information has to be 'deactivated' and protected from either being the subject of, or the object of causation. This deactivation is achieved by embodying it in a form that makes no direct sense - in an encrypted form - it is effectively meaningless and can only be acted upon after it is transcribed (decrypted) into a meaningful 'active form'. A piece of commercial software could be like this: the data you receive is meaningless, uninformative, until you provide the activation key, which enables it to be decrypted and turned into the active form of a functional program. The decrypted 'active form' of DNA is transfer RNA (though that is a gross simplification of course).
Having protected DNA, the issue becomes one of which parts of the information string to transcribe and when. The instructions for this are themselves embodied in the DNA, which for the same reason are also inactive until they can be transcribed. DNA cannot do anything other than as part of a large and well coordinated system of molecular parts which coordinate the transcription and translation of the right bits of information at the right time. The elaborate system of self regulation found at the heart of every cell has all the appearance of a computer algorithm running on molecules.
A great deal of the DNA sequence is actually devoted to controlling its transcription and translation. In total, surprising little is used for specifying the sequence of amino acids in proteins to be fabricated. A lot of the rest is the control algorithm and the basic unit of that is the binding of transcription controlling proteins to turn the reading of genes on or off. These proteins have a surface that (approximately) fits a particular 'motif' - a small sequence in DNA, so they recognise it when they fit (this is termed binding, and a bit like a key turning a door lock). They don't usually fit exactly, so are able to bind to a rang of similar sequence motifs with different affinities.
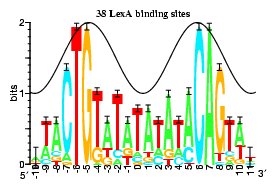
Using the basic idea of entropy, we can quantify the probability of matching a particular nucleotide in the sequence, and of course, the inverse of entropy is the amount of information (in bits) that is conveyed by that point in the sequence. The diagram illustrating this theme (on the right) is an example of how we convey this idea (it is a sequence logo, attributed to Tom Schneider). The logo represents the motif that the gene-regulatory protein is supposed to fit and it helps molecular biologists work out which genes it is most likely to affect. At each position along the DNA chain, there are four nucleotide options (C,T,A,G) and the frequency with which each occurs within the motif is represented by the size of the letter (see they are stacked at each position). With this, we can see that where there is only one letter (e.g. position 6) it is huge and the information embodied there is the maximum of 2 bits (i.e. at that position every sequence the protein binds to has the same nucleotide in common). At positions -10 and 11, we see a stack of all four tiny letters - they all occur with similar frequency, conveying approximately no information at all (among all the motifs it binds to, there is no consistency at all in the nucleotide appearing at that point). It is worth looking at the large and well maintained library of transcription binding profiles with their sequence logos and the statistics behind at JASPAR (a very accessible site). It is now apparent that many diseases have their roots in faults with these gene-regulatory sequences.
Computing in DNA
DNA and RNA embody algorithmic instructions, many of which direct actions upon the molecular information itself, particularly in the broad category of regulatory systems in which the function of many genes is to switch on or off, or to modify the transcription or translation of other genes (even including each other). These regulatory instructions form complicated networks that respond to events, both intra- and extra- cellular. Collectively they form a computer algorithm: the computer is the set of molecules involved, the algorithm being the logical outcome of their interactions.
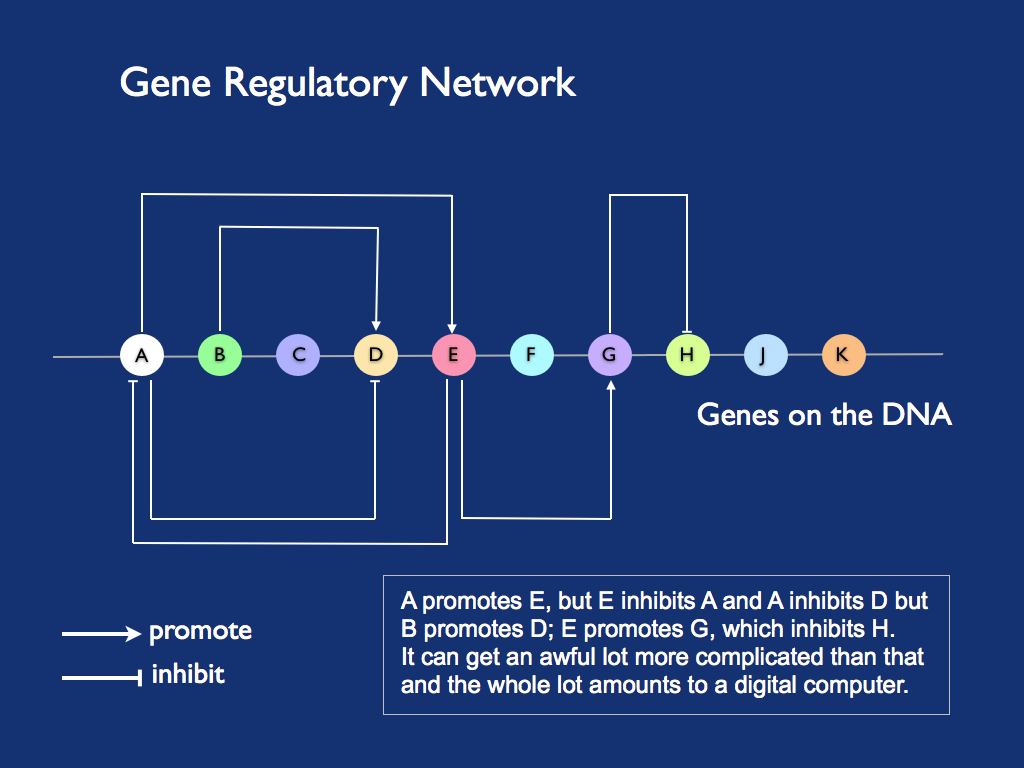
Gene regulatory networks (illustrated above) are deduced from gene expression data - observing the proteins that result from transcription and translation of genes. Quite a few of those proteins are regulators themselves (e.g. the 'transcription factors' mentioned above in the context of sequence logos) - their role is to either promote or inhibit the expression of other genes (actually some proteins inhibit their own making, in a classic example of negative feedback). The details of all this can be found in any biology textbook, here we want to concentrate on the information content and the way it is processed. Most strikingly, and different from everything else biological, DNA and RNA information is digital (more precisely, it is discrete). This was recognised by Sara Walker and Paul Davies in their (2013) description of the information systems of life as digital or analogue, associating the former with replication and the latter with metabolism. For this reason, gene regulation networks are often referred to as 'genetic logic' (the term 'genetic algorithm' was already taken). [Making a page on gene regulation networks is on my to-do list].
Molecular signalling
As well as that, It is now appreciated that many of the cell’s processes constitute communication within and among cells, via cell signaling. Molecular messengers carry information about the state of the environment, both internal and external and they communicate this with receptor molecules that in turn trigger cascades of molecular-based responses in the cell. This cell signaling enables coordination among bacteria (quorum sensing) and coordination among cells within a multi-cellular organism, indeed without it multicellularity would be impossible. It also enables a cell to adapt to its environment and to manage its own homeostasis and the coordination of complex processes such as reproduction, in which several different structures have to act in the right way at the right moment. The combination of genes, genetic regulation networks and intracellular signaling systems is collectively the cellular operating system (COS) and (just as in a computer) it is organised as a nested hierarchy of subsystems within systems (illustrated below). However, unlike the digital computer it comprises a mix of digital and analogue elements with a large range of specialised components (e.g. thousands of enzymes). The computer on your lap, or in your phone is an inert lump of metal and plastic (plus a few exotics like doped silicon) - without its operating system it cannot do anything at all and in fact it is not really a computer in any meaningful sense without the operating system, which it maintains as 'pure' information in memory embodied by variations of electrical charge. A biological cell is no different in that respect - without the COS, which is embodied information, it is just a blob of wet chemicals and cannot do anything, especially not the very thing that defines it as a biological cell - to live.
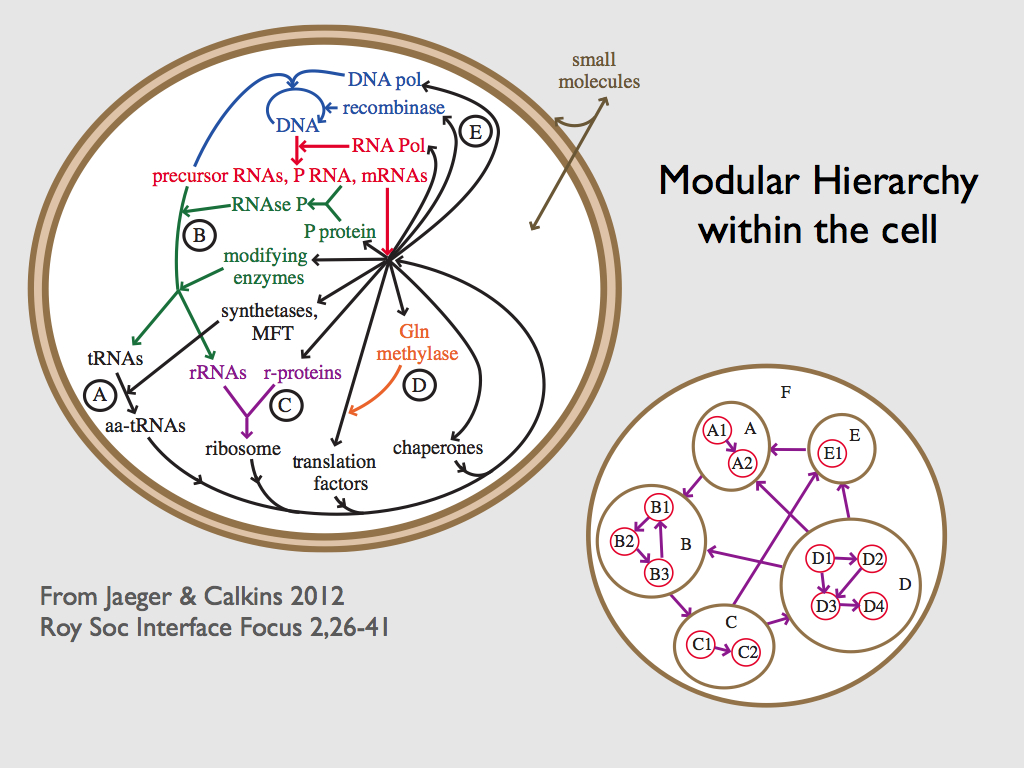
The COS is the information that coordinates the thousands of chemical reactions that collectively constitute life. Its overall effect is to be the formal cause of autopoiesis - the continual self-making of the cell.
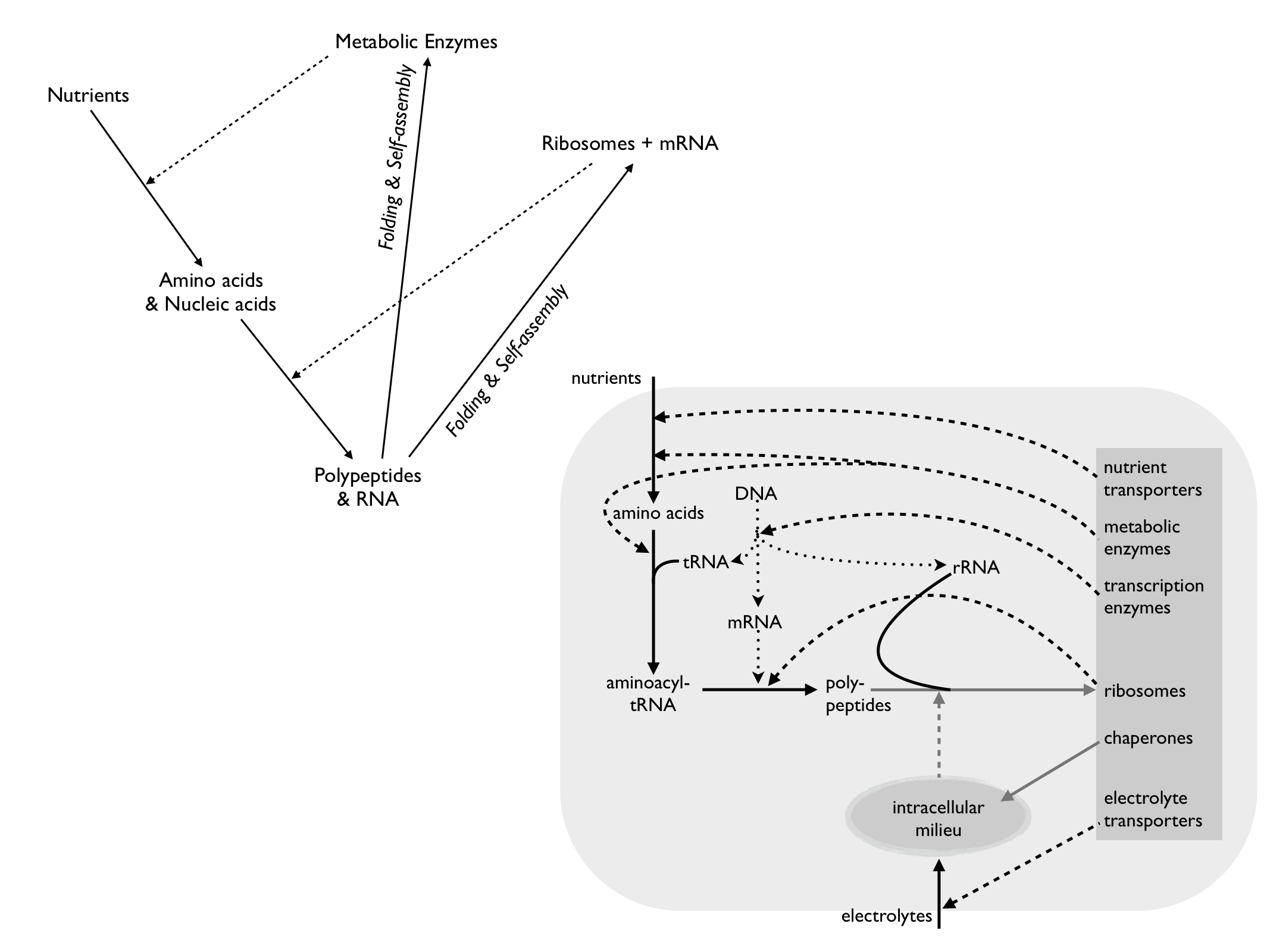
The diagram above shows two levels of abstraction of the COS, both by Prof Jan-Hendrik Hoffmeyr of the University of Stellenbosch (though he does not call them COS). The top left was published in 2007 and emphasises (at the highest level of abstraction) how the COS results in closure of efficient causation. The one on the lower right (redrawn by me as a copy of his Fig. 7 in Hofmeyr (2017)) shows the broad classes of chemicals involved and how they interact to produce a meaningful 'whole', that is a living system. As in the top left diagram, solid arrows represent material causation (e.g. chemical transformations) and dashed arrows show efficient causation (e.g. catalysis). Dotted arrows add to these to indicate formal cause by sequence information (functional code). All this talk of causation is underpinned by the fact that cause is the result of physical forces (in this case the forces of molecular interactions) being constrained by embodied information and is explained further on the pages about the physical basis of cause, code biology and circular causation. Hofmeyr (2017) emphasises that, by providing an environment in which peptide folding (tertiary structure) leads to functional forms of proteins, especially enzymes and transporters (grey box), the intracellular milieu (especially including chaperone molecules) acts as an efficient cause in its own right. The grey arrows indicate supramolecular processes and what they depend on, e.g. the efficient cause of folding RNA and polypeptides into functional ribosomes. This biochemical overview demonstrates the property of closure to efficient causation as well, if you follow the arrows and know what they mean. His theory was updated and eleborated upon to produce his 'Biochemically-realisable relational model of the cell' (Hofmeyr 2021), explained on our blogpost, and it features prominantly in my explanation (Farnsworth, 2022) of biological causation, using the ATP synthase complex as an example.
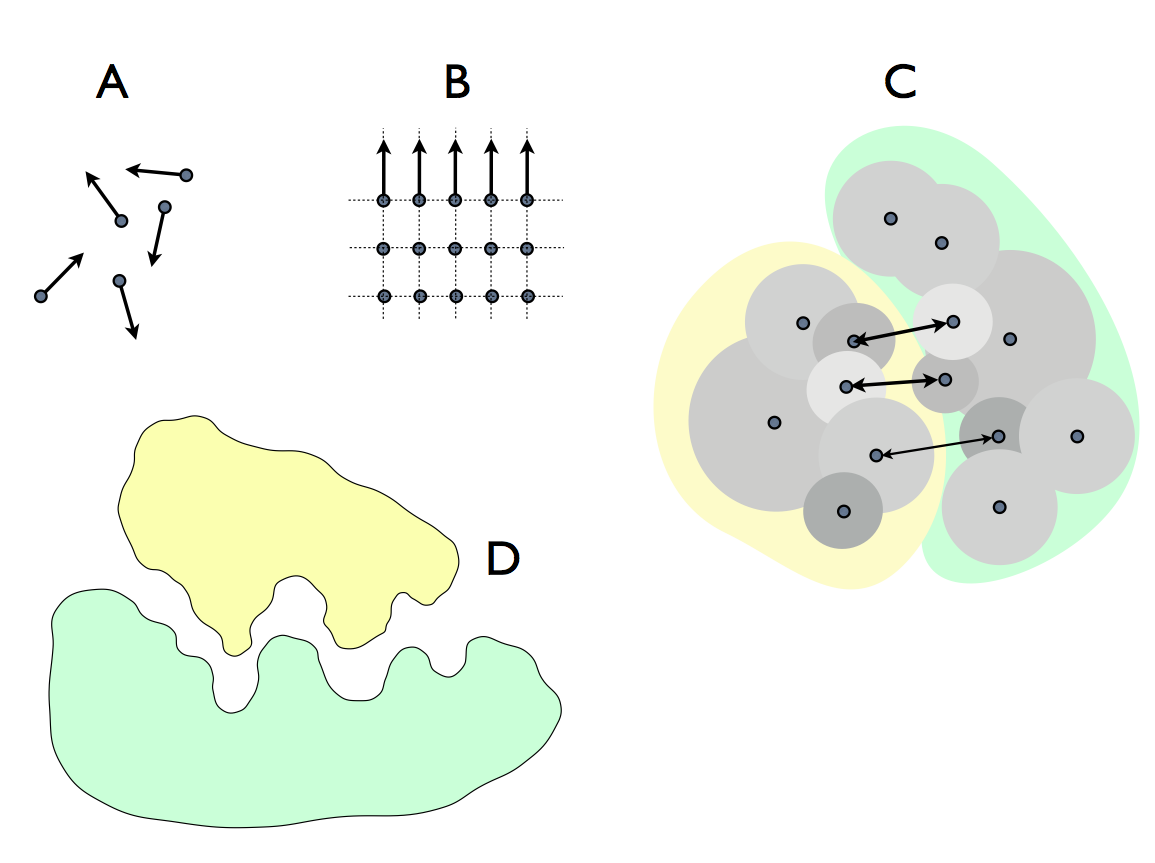
We have seen that molecules themselves embody information in their form. That was obvious in the linear chain of a nucleotides forming a nucleic acid : G U A C G A ... it even looks like writing put that way. But more generally, every molecule is a persistent pattern of atoms in space and as such it embodies information as the pattern of locations of the atoms. In the absense of life, its persistence is brought about by thermodynamic gradients - it minimises local Gibbs free energy. Life is unique in adding (greatly) to that by fabricating molecules, piecing them together in specific configurations (usually this requires enzyme activity and the special chemical conditions maintained by the cell). In fabricating a molecule, information is assembled and stored. If the molecule is a functional protein, there is at least one part of it (the active site) that has a shape which functions, like a tool, in cellular biochemistry. This shape is iteself embodied information and it works by interacting with other molecular shapes to bind, distort, move and cleave other molecules: function. In the diagram below (from Farnsworth, 2022), you can see A) atoms moving at random (no information); B) atoms arrayed in a stable (energy minimising) crystal (rather little, but some information - think of compressing the data file that describes all the locations); C) two molecules interacting through the electrical forcefield generated by their atomic electron shells; D) this idea represented in application as a signal molecule (ligand) fitting its receptor.
A single protein molecule can, in many cases, be folded up in more than one way and can reliably switch between each way, trigged by e.g. phosphorphorylation. This conformational switching is another rather obvious way to carry and transmit information (and it is the basis of a lot of intra- and inter- cell signaling). Where there are switches, there is the possibility of computation (remember that electronic computers are made from millions of silicon switches). Long range inter-cellular signaling is achieved by hormones in multicellular organisms. The hormone, e.g. insulin or serotonin or adrenaline (epinephrine for the North Americans) is a very particular molecular shape - one that matches a receptor for it in, for example among the G-protein coupled adrenoceptors. In this case the specific hormone adrenaline is a ligand for the binding site on the adrenceptor molecule and if a molecule of adrenaline were to float by close enough, it would bind onto the receptor site and this would in turn cause the adrenoceptor to change its shape (as a molecular switch) and thereby trigger changes in the cell in whose membrane it is embedded. The receptor is very discriminatory (though in this case adrenoreceptors turn out to respond to mimics such as the beta blockers used as hypertension medication (heart pills). That means, if we remember Shannon's information theory, that it carries information in the senseer possibilities: the coupling of hormone and receptor is a strong constraint, so reduces entropy and thereby constitutes information. More obviously, the hormone works as a signal to say we need some action here! (and I'm sure we all know the feeling of adrenaline). Just as the arrangement of flags in a semaphore symbol is a particular configuration, so the outer surface shape of the hormone is a particular configuration that carries an unmistakable message. It is just one molecule, it is either bound to the receptor or not, so it is just a binary bit of information. Still, it is the shape that matters and that shape is embodied information - the information needed to construct the molecular form of the hormone and the information it embodies in its form. The information embodied by molecules can be worked out from their geometry (or topology), as Rashevsky (Robert Rosen's mentor) showed in 1955. The information embodied in the form of nucleotides was found by Sakar et al. (1978) and a host of other molecules, by Bonchev (1979). 1955 was obviously a good year for this sort of thing because it was then that Morowitz estimated the information content (embodiment) of an E.coli bacterium as 4.6x10^10 bits (which is about 180 megabytes and I think that puts a tiny 'simple' organism in perspective).
The most fabulous example of cellular coordination by inter-cell signaling, is that of the mammalian immune system. Not only do cells send a multitude of messages to each other and act upon these messages, the highly complex network of their communications itself amounts to a computer. What is it computing? Well faced with a bewildering array of different molecules, it sorts those belonging to the self from those that are foreign and selectively directs waves of attack from multiple destructive systems to destroy the bacteria, viruses, parasites and bits and pieces of them, as well as bits of the body it belongs to (dead cells and cancer cells) in a clean up operation that would keep a digital computer busy for a very long time.
Autopoiesis (of course!)
The computer on your desk (or in your phone) has a power supply and a lot of information processing hardware. You could say it has a physiological system (the power conditioning, distribution and management system) and the main business: an information processing system, consisting of memory, processor chips and some other parts. The cell has both of these systems too, but crucially it has a third processing systems that no computer, nor any technological object has. This system is the one which continually makes the cell from within (a by-product of which is reproduction). It is the most obvious difference between something that is alive and something that is not; between an organism and a machine. This self-making (autopoietic) system is also an information processing system because what it does is arrange the correct materials (molecules) in the right place at the right time. It is responsible for creating and maintaining embodied information. How it does this from the beginning of its life is a matter of information bootstrapping - the 'booting up' of the cellular operating system.
This explicit information processing system of the cell takes care of the routine management of homeostasis, responding to changes in the external environment and communications from other cells as well as the translation of information from DNA into functional forms as proteins or whole-scale reproduction. It is made up of the gene regulation networks and the cell signalling pathways and it interacts with and controlls the cellular physiology. The physiological system of the cell is also a kind of information processor (often referred to as analogue) because it sorts the desired molecules from those that should be got rid of, it regulates energy acquisition and consumption and generates the raw materials for autopoiesis, whilst itself being a product of that same autopoiesis (as explained in Hoffmeyr's diagrams above). All the actions of living performed by a cell are examples of information processing - computation - implemented through biochemistry (see Bray 2009).
Molecular Biology makes use of information theory
(This part is still very much in draft)
More narrowly, Shannon’s definition of ‘information’ as a decrease in the uncertainty of a receiver has enabled quantitative analysis of biomolecular systems, using concepts such as ‘mutual information'* and ‘channel capacity’*. These aspects of information theory have allowed the development of a straightforward and practical method of measuring information in genetic control systems. This enables us to answer questions such as: How do genetic systems gain information by evolutionary processes?
Tom Shneider's paper here explains and uses the method to observe information gain in the binding sites for an artificial protein in a computer simulation of evolution (there is a list of Tom's paper's here). The simulation begins with zero information and, as in naturally occurring genetic systems, the information measured in the fully evolved binding sites is close to that needed to locate the sites in the genome.
* Definitions available in the Glossary for Bio-molecular Information Theory
by Tom Schneider and Karen Lewis
References and Further Reading
Adami, C. Information theory in molecular biology. Phys. Life Rev. 2004, 1, 3–22.
Adami C. 2016 What is information? Phil. Trans. R. Soc. A 374: 20150230. http://dx.doi.org/10.1098/rsta.2015.0230Adami C. 2016 What is information? Phil. Trans. R. Soc. A 374: 20150230. http://dx.doi.org/10.1098/rsta.2015.0230Adami C. 2016 What is information? Phil. Trans. R. Soc. A 374: 20150230. http://dx.doi.org/10.1098/rsta.2015.0230
Bonchev, D., 1979. Information indices for atoms and molecules. MATCH 7, 65–113.
Bray, D. Wetware: a computer in every living cell. (2009). Yale University Press, New Haven, USA.
Hofmeyr, J.H.S. (2007). Systems biology: philosophical foundations.. Elsevier, Amster- dam.. chapter The biochemical factory that autonomously fabricates itself: a systems biological view of the living cell. Systems biology: philosophical foundations. pp. pp 217–242.
Hofmeyr, J.H.S., (2017). Handbook of Anticipation: Theoretical and Applied Aspects of the use of Future in Decision Making. Springer. chapter Basic Biological Anticipation. 11, pp. 219–233.
Hofmayr, J-H. S. (2021) A biochemically-realisable relational model of the self manufacturing cell. Biosystems. 207:104463. doi:10.1016/j.biosystems.2021.104463
Farnsworth, K. D., Nelson, J., Gershenson, C. (2013). Living is Information Processing: From Molecules to Global Systems. Acta Biotheor. 62: 203-222.
Farnsworth, K. D. (2022). How an information perspective helps overcome the challenge of biology to physics. Biosystems. 104683 doi:10.1016/j.biosystems.2022.104683.
Goodsell, D. S. (2009). The Machinery of Life. Springer, New York.
Morowitz, H., 1955. Some order-disorder considerations in living systems. Bull. Math. Biophys. 17, 81–86 doi:10.1007/BF02477985.
Rashevsky, N. (1955). Life, information theory, and topology. Bull. Math. Biophys. 11, 229–235.
Sarkar, R., , Roy, A., Sarkar, P. (1978). Topological information content of genetic molecules - I. Math. Biosciences. 39, 299–312.
Walker, S.I. and Davies, P.W. (2013). The algorithmic origins of life. J. R. Soc. Interface R. Soc. 10, 20120869. [1207.4803]
Molecules build the patterns necessary for life
One of the core principles of our particular understanding of life is that it is constructed from a nested hierarchy of informational structures (patterns), each level creating the next above by self-assembly. The lowest level of relevance to biology is that of molecules, but understanding how a collection of different chemical 'species' can eventually lead to a living organims requires an appreciation of how physical forces, combined with quantum rules, create the molecules and how these assemble into supra-molecular structures and these in turn form functional complexes. Since information is embodied in the particular arrangement of an assembly of parts, all molecules embody information (as a particular arrangement of atoms). Molecules can only interact in a limited set of ways, specified by their shapes - this limitation also embodies information. Networks of interactions among a host of molecules embody information because they are each structured in a particualar way (which molecule is connected to which others and by which reactions). Sets of such networks are combined in particular ways to make functional complexes. These perform the functions of the cellular operating system.We strongly recommend David Goodsell's book and its wonderful illustrations as an aid to appreciating the molecular machinery of the living cell. In fact, the IFB project hopes to engage its author David Goodsell in future developments.
You may like to read our 'tutorial' paper here: How much information does DNA instantiate?
This Theme aims to:
- Develop an information-based understanding of living processes at the molecular level;
- Apply this to better understand how life arose, evolved and builds complexity;