Molecular Codes
Keith Farnsworth 2020 (revised July
2022).Also see my lecture on Code Biology and Autonomy.
Molecular code immediately brings to mind the DNA (and RNA) code of course, but there is much more to it than that, as leading proponent of Code Biology Marcello Barbieri shows with his (several) books and journal publications.
First what do we mean by code?
The usual illustration of a code is the Morse code. It has the primary attributes of a relational code as defined by the Society of Code Biology:
A code is a "set of rules that create a correspondence between two independent worlds". This is just one of several meanings of code in broader usage, but it is the one that lies at the heart of code biology and is applicable to what Barbieri and followers term 'organic codes'.
The correspondence referred to is a mapping from one set of symbols to another (confusingly, each set of symbols can be termed a 'code' in computer science). More formally, if A and B are alphabets, then a mapping between the members - A mapsto B - is what I am calling a relational code.
So with Morse code, the Latin alphabet A...Z (plus a few additions) is mapped onto a set of symbols made from sequences of dashes and dots. Also in semaphore (waving flags) the Latin alphabet is mapped to a set of different combinations of angles of the two flags and in maritime flag code, to a set different flags. In every case, the mapping is one to one (unique element).
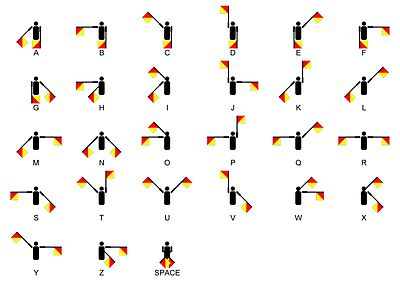
Fig 1. semaphore code from wikibooks.org/wiki/Scouting/BSA/Signs,_Signals_and_Codes_Merit_Badge
where there are more codes to see (e.g. braille).
Not included in the definition above, but also important for code biology, is the arbitrariness of the relation between the two alphabets. There is no necessary connection between any letter of the alphabet and any Morse or semaphore representation of it. Given that, the mapping is in fact a set of elemental mappings - a mapsto b - one of these for every pair of corresponding members of the alphabets.
The two alphabets being mapped are the two worlds referred to in code biology, but what is really meant is two independent sets of molecular species that can, by presence and absence, sign a state. This is most obvious and best known in the mapping between nucleotide sequence (strictly codon sequence) and amino acid sequence (together making the the genetic code). It is not immediately clear that codons are unrelated to amino acids (i.e. that the code is arbitrary), but a considerable body of experimental (biochemistry) evidence now shows that it is (references in Barbieri (2018)). Succinctly, any codon can, in principle, map to any amino acid. The fact that there is a specific and almost unique mapping from each codon to each amino acid indicates an information rich translation system. Such systems (the rules for mapping) are the essence of every code.
Barbieri (2003, 2018) argues that there are many relational codes in biological systems and that the emergence of each has introduced a great innovation - a major evolutionary step, such as the development of eukaryotes and also of multicellular organisms.
Of immediate interest here is the question of how the arbitrary mapping is maintained in a real system, such as a cell. The answer must be that there is some other (translational) system that 'knows' the mappings of all the elements, i.e. there is some system that embodies the information needed to specify the mappings.
Genetic code: translation by shape matching
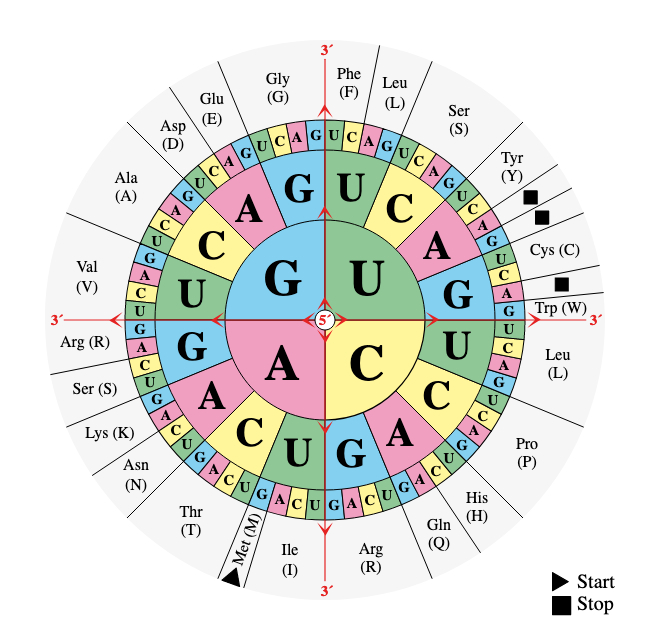
FIG. 2. The genetic code wheel - from Wiki Commons
see here for explanation and help with reading it.
Let us revise: in protein translation, there are 20 standard (proteinogenic) amino acids to map to (that is one alphabet) and 4^3 = 64 codons (triplets of nucleotides) to map from (we could add some punctuation marks such as stop codons and non-standard amino acids for completeness). The mapping is all done physically, with molecular shape matching of course. To go from one to the other, we must have a translation system: something that, given a particular codon triplet, selects a specific amino acid and presents it to the ribosome for adding to the growing peptide chain. That is exactly what we find in every cell capable of protein synthesis (see Figure 3). There is a set of (23 known, including two for lysine) enzyme molecules called aminoacyl-tRNA synthetases (aaRS for short), each member of which has a particular pair of shaped 'slots': one that binds the right amino acid and the other that binds the transfer RNA (tRNA) molecule that corresponds to the right codon. Their name describes something of what they do (join amino acids to tRNAs, making aminoacyl-tRNA products). The set of aaRSs are a sort of matching tool (imagine you had a mechanics socket set and a pile on nuts of unknown sizes - you could identify the nuts by seeing which socket driver they each fit). The tRNAs are represented by the nuts and each is specific for a particular proteinogenic amino acid and also for a particular codon in the mRNA (which it achieves by having a region that 'recognises' the codon by base-pairing. There is a degree of 'belt and braces' about the code translation here, because a particular tRNA only matches one amino acid and recognises only one codon.
tRNAs are themselves intermediaries (adapter molecules) that match one particular codon each. In fact there are about 60 different tRNAs in bacterial cells and 100-110 in mammalian cells, whilst there are 64 possible codons. The tRNA can (covalently) bind to only one particular amino acid, but because several codon sequences typically specify that particular amino acid, there is a set of several tRNAs that bind to it. In effect the tRNAs embody the genetic code as a translation system.
Since a particular tRNA can only sit in a particular aaRS molecule, which in turn can only connect it to a particular amino acid (the one it can bind to in its other slot), the aaRS molecules effectively match codons to amino acids, performing the code translation and thereby 'double check' the matching of amino acid to codon. But these aaRS molecules are a set of forms, each a choice and each embodying information, specifically in this case the shape which holds only one kind of amino acid and one (corresponding) tRNA. The set of aaRS enzyme forms is internally embodied information and so is the set of tRNA molecules.
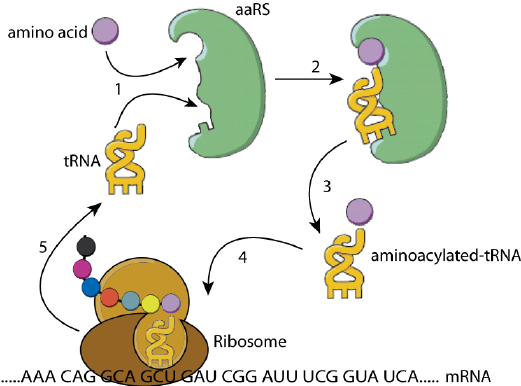
Fig.
3. The protein translation system using aminoacyl-tRNA synthatase
(aaRS) molecules to match the right amino acid with a particular codon
that is represented by its complementary tRNA molecule. The set of
aaRS molecules and corresponding tRNA molecules form a code
translation system that works by the selective binding of molecules,
strictly following their forms: it is embodied information. Figure
source: Wang et al. (2012) , Fig 1.
This might surprise, but the preserved choices that define the code in the embodied information of tRNA and aaRS shapes is one of the deep foundations of freedom and ultimately of free will. These molecules could be a wide range of shapes, even many potentially functional shapes in their binding sites: there is nothing chemically or thermodynamically inevitable about the shapes they actually are within cells. There are rather few examples of such thermodynamic indifference beyond what we see in living systems and those we do find are very simple and offer few choices - typically just two (as in stereochemistry). But life absolutely abounds with complicated, wide ranging, thermodynamic indifference, where the outcome of a chemical reaction is so loosely determined that it requires the action of a catalyst (often a network of interacting proteins) to establish a particular outcome. In life, this particular outcome is consistently produced because the additional information needed to constrain the range of options (left by thermodynamic indifference) is preserved by the act of living: specifically, it is embodied in the catalytic molecule complex that informs the reaction.
Out of thermodynamic indifference comes arbitrariness and out of embodied information constraint comes choice.
More Biological Codes and symbols
It does not end there. Barbieri identifies several more biological codes in his 2018 paper, listing the metabolic code (first described by Tomkins (1975), the sequence codes, histone code, sugar code, splicing codes, the compartment code and the cytoskeleton and tubulin codes and more at the cellular level, together with the Hox code for body plan development and yet more - for apoptosis, and intercellular communication etc..
But not all of these match the definition of a mapping between two independent domains (that's domain and codomain if you are a mathematician), achieved by a third matching (adapter) domain that translates between them; the translation being arbitrary, i.e. not determined by any chemical or logical connection between the two domains. Within and among cells, adapters should be molecules whose shape can be recognised (i.e. it correlates with other molecules) in both the domains it connects. That is certainly true of tRNAs and also for the effector proteins in post-translational modification (PTMs) of DNA associated with histones (the proteins making up nucleosomes in eukaryotic organisms), convincingly demonstrated by Kühn and Hofmeyr (2014). Both genetic code and this histone code connect the world of DNA (pure - formal cause - information) to the world of functional (efficient cause) proteins.
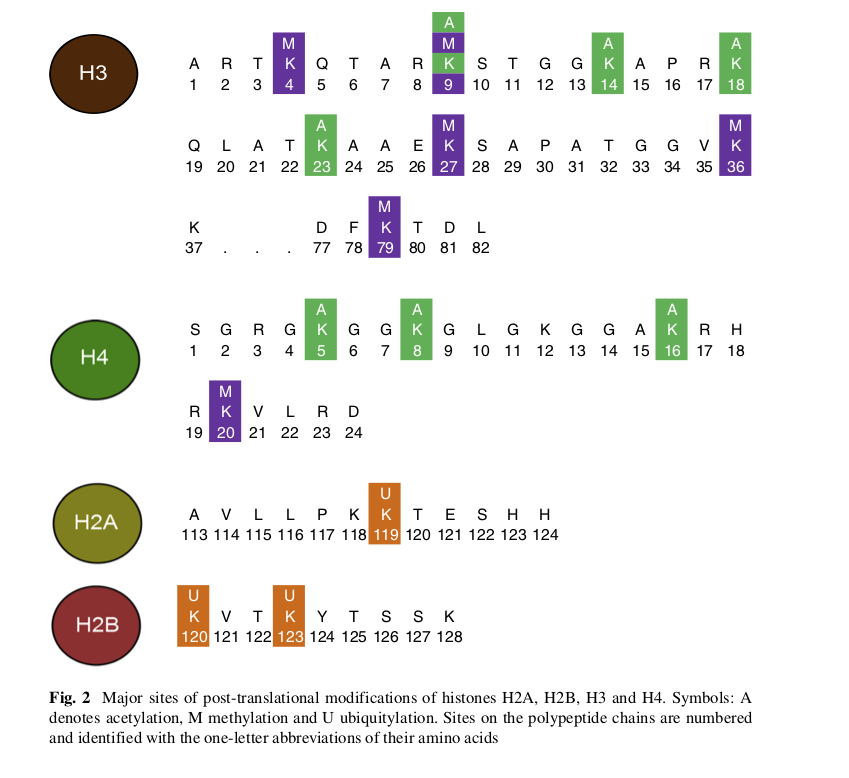
FIG. 4. The histone code - taken from Kühn and Hofmeyr (2014) Fig.2 with original caption.
This might not be the complete code, the authors concentrated on the PTMs for which sufficient evidence was available at the time. The circled symbols down the left hand side are the histones themselves, numbers indicate the position on the histone polypeptide, with letter symbols for the amino acids (residues) at those locations (e.g. K stands for lysine). The colours match the modifications of those residues : acetylation in green, methylation in purple and ubiquitylation in orange. This action takes place on the N-terminal tails of the histones, which appear as wavy prongs sticking out of the nucleosomes.
The histone code is still usually described as hypothesis, but it is now supported by a lot of evidence and the idea has found applications in many explanations of diseases and fundamental biological phenomena. Essentially, it provides an addition to the genetic code by controlling gene expression (in conjunction with other sub-systems) and therefore contributes to epigenetics, often described as an additional cellular memory. Given the very large number of modification states for each histone and strong interactions among them, it is still bewilderingly complicated, so the picture above serves only as an introduction. Still, the point here is that the histone code fulfills all the requirements for a genuine 'organic code'.
The situation for the metabolic code is rather different. Here, signaling molecules, especially cyclic adenosne monophosphate (cyclic AMP) indicate a looming crisis to which the cell has a response. Cyclic AMP signals carbon (energy) starvation and it is noted that cyclic AMP is not chemically related to the molecules in energy metabolism: it is an arbitrary signal (Tomkins (1975) speculated that it may have acquired this role in early organisms when a "primordial kinase", at one time a catalyst of phosphorylation of glucose, was "idling" in the absence of sufficient glucose molecules). Another apparently arbitrary signaling molecule - guanosine 5'-diphosphate 3'-diphosphate (ppGpp) indicates nitrogen, especially amino acid, starvation. Both cyclic AMP and ppGpp act as effector molecules, triggering a series of changes in the cell, interacting with DNA and metabolic pathway switches to mount an emergency response. However, whilst good arbitrary signals they may be, they are not adapter molecules connecting one domain of information embodiment with another. For that reason, I would prefer to call the metabolic code a metabolic signal complex. The signal molecules can be recognised by an arbitrary set of control systems (e.g. metabolic pathway switches and gene-regulators), but do not specify a mapping between one set of symbols and another. Indeed they are symbols, quite independently.
Apart from the genetic code, the most obvious case of an organic code that fits the definition is that used in cell signaling, which I say a little more about below (Codes as Signage systems). For intracellular communications, the transmembrane transducers, usually protein complexes, act as adapters linking the world of extracellular chemical messengers, typically hormones, to the world intracellular control circuits. In principle any extracellular signaling molecule can be matched with any intracellular signaling molecule: the adapters all have a binding site for each: a receptor on their outside end and a mechanism for releasing an internal messaging molecule on the inside end. Along with others, the signal transduction codes are described in Barbieri (1998, 2003).
Codes as Freedom
This idea has been written up as a journal publication
- see How biological codes break causal chains to enable autonomy for organisms
K. D. Farnsworth (2023) BioSystems, 232. doi:10.1016/j.biosystems.2023.105013
Freedom to act and freedom to choose are uniquely features of all living things (see our pages on causation and on autonomy). Freedom means release from the physical chain of cause and effect that determines all that happens in the universe. The only way to avoid that causal determinism is to be closed to efficient causation and that implies having internal physical information to call upon. Closure to efficient causation implies an ontological and causal difference between what is internal and what is external. Information constrains forces to produce cause (see the physical explanation of cause). Freedom results from internal information being able to constrain forces and thereby create internally sourced cause. This in turn gives any system that is closed to efficient causation the attribute of agency.
But the form of molecules is usually determined by thermodynamic chemical necessities, it is in fact inevitable, so there is no clear escape from the causal determinism of the universe at this point. However, uniquely as far as we know, life creates and needs molecules that are not chemically inevitable. In fact, they are fabricated by living processes. For example (and it is an important example) proteins are functional only in their correctly folded form (i.e. if they have the correct secondary and tertiary structure). This structure is thermodynamically stable, but rarely inevitable because usually there are many thermodynamically stable folded structures that the polypeptide could fold into (the functional one can be thought of as a local minimum in free energy, rather than the global minimum). Almost all proteins in a cell are therefore a choice - each is one chosen form of several possible forms. Given that, its form is information in two respects. First the shape itself requires information to describe (or construct) it, so it embodies information that way. It is also one out of several possibilities, so its existence is in fact information in the Shannon sense of a selection or constraint on the range of possibilities.
The aaRS - tRNA system described above is a very good example of this. In principle it is possible to make an aaRS molecule with any pair of slots so that it could match any amino acid to any codon. Not only that, but there is no chemical necessity for the anticodon end of a tRNA molecule to be restricted to those appropriate for the amino acid that selectively binds at the other (free) end. This is the basis of the arbitrariness of the genetic code.
If there are 20 amino acids and 64 codons, that would be 64x20 = 1280 possible pairings, but in fact, cells have only 20 pairings (roughly speaking - there are a few little exceptions). This constraint to 20 pairings, from all possible, is information. It is the information that specifies the (standard) genetic code for a cell. As part of the cell and made by the cell (it constructs the aaRS molecules), it is internal to the cell - indeed it is part of the causal loop that collectively constitutes closure to efficient causation.
Jannie Hofmeyr (2007; 2017; 2018) was perhaps the first to state in the scientific literature that this opportunity to embody information in a system that is closed to efficient causation arises from the fact that biomolecules such as proteins are fabricated, not simply the result of chemical reactions. They are actively constructed, requiring information to make them as they are. This information comes in the form of an mRNA template that specifies the order in which amino acids are strung together (primary structure) and also in the form of folding of the resultant polypeptide string (secondary and tertiary structure). It is this folding that gives the protein its function (e.g. as a catalyst) and it is the collective action of catalytic proteins that determines the realised, functional, form of it. That functional form is determined by internally embodied information which informs (constrains) the action of non-covalent atomic forces to produce the functional result. This is only possible by the action of particular functional molecules constraining the force fields around the atoms in the fabricated protein by the action of their own atomic forces that were themselves constrained (informed) by that same process (see physical explanation of cause). In other words constrained forces are both required for and produce the systems that constrains forces in such a way that they can constrain forces in that way. Constraining forces to produce a particular outcome from a range of several possible is what we term efficient cause. The circularity of an efficient cause being necessary to give rise to the very same efficient cause that it gives rise to - that circularity is the closure to efficient causation.
The fabrication of functional molecules, freeing them of chemical necessity, is the elemental step in creating freedom and choice, from which agency and autonomy emerge. So far, life alone is known to have the facility to fabricate molecules. To achieve it, evidently a system must itself be constructed from fabricated molecules - and so we come to the familiar 'chicken and egg' problem that lurks within all serious questions about life. As always, the answer to the riddle is bootstrapping and in this case, it involves the early development of a translational system for making proteins that long preceded the last common ancestor (LUCA) of all known life. How the first translation system, along with the first genetic code (they had to develop concurrently) emerged is still a matter of speculation because so little evidence survives the takeover of life by the modern, highly developed form it now takes in all known life. (It is worth commenting that alien life, should we ever find it, might well be entirely different at this most fundamental level and if it is not, that will be a big clue as to how life comes into existence at all).
So the freedom that coding affords an organism is the freedom from external cause. It works in transducers (e.g. chemoreceptors embedded in the cell's outer membrane), where cause and effect are transformed into stimulus and response. It also enables the fabrication of components needed for life, free from chemical necessity. The code is information that is embodied as long as the cell is alive. It is embodied separately from the molecules taking part in the process of living, but strongly influences what they do. To that extent it acts almost as an information catalyst, though it does not accelerate reactions (as true catalysts do). Instead it allows a template molecule to select those reactions that are required and prevent the others. This behaviour is only possible if the template molecule remains unaffected by all the reactions taking place, other than those that 'read' its information.
The code is the way template molecules (RNA and DNA) are protected from those other reactions. It is as if the active chemistry of the cell were conducted in a language that it cannot understand and vice versa. To illustrate that, imagine you are a karate instructor in the following (rather odd) situation. Four trainees stand an arm length apart, each at one of the four compass points: N, S, E, W, with you standing in the middle. You have a card of instructions for them to follow. When you look at it, you see the first instruction is punch to the left. Clearly there is a problem: you are going to get your right ear bashed. The next instruction is kick forwards - that could be even worse as South slams her foot into your back. One possible answer to this conundrum arises from the fact that each of the trainees speaks a different language as well as your common language. Let's say North speaks Russian, South, Portuguese, East, Arabic and West speaks Irish. Now you can safely shout the first instruction (punch to the left) in Irish - nobody gets hurt. You can issue the second in Russian and you can say "raise your arm to defend from above" in your common language (let's say Japanese). In every case, you are neutralising the effect of the command on yourself by encoding it in a language that only the intended recipient will understand and act upon. To make that possible, you and each of the trainees must hold a lot of information that is not the instructions - it is, of course, the languages. Those languages are codes: the actions performed are arbitrarily associated with the sounds we make when we speak words. We know the association is arbitrary because the sounds are different in each language.
There is something else about languages that displays the freedom accorded by codes. We can assemble any (real) words together to make arbitrarily complicated meanings and there is an effectively limitless set of possible meanings we can convey with groups of words. So it is with codons on DNA and RNA. There has to be an 'agreement' or standardisation of what each means (that's what the genetic code is for), but from then on, any concept, i.e. any polypeptide, can be fabricated following the particular assembly of codons decided upon. At this molecular level, the genetic code is crucial to releasing biochemistry from the constraints of chemical necessity - physical cause and effect.
Codes as Signage systems
Another (related) special feature of life is the use of signs and signals (the study of which is semiotics). The nucleotides in an RNA molecule are each signs and the linear sequence of them is a signal. Signs and signals convey information and that is all - they do not physically determine what happens next; instead they can only be responded to (and perhaps will not be). It is the feature of codes as 'arbitrary' that makes them able to function as signage systems. The thing about signs is that on their own they have no meaning and no effect. They are only effective if they can mean something to a receiver system. It decides what to do (or not) about them when it detects them.
This seems obvious when we are thinking of human-designed technical systems. In those, signs indicate states of the environment to which the system should respond, for example a stop sign for a driverless car. The important point here is that the stop signal does not stop the car, it only provides information about the environment for the car (or its driver if there is one) to respond to. That response remains an option because it is not physically related to the stop sign at all. We could reprogram the car to take a red (stop sign) to mean engage reverse gear (and cause a lot of accidents). This lack of physical connection is what is meant by the difference between 'cause and effect' on the one hand and 'signal and response' on the other. Signal and response is only possible, only meaningful, if the system making the response is autonomous, i.e. it is free from the determinism of cause and effect. The only things that are free like that are living systems (and the technological artifacts that we make to be). So the very idea of a sign or signal is itself contingent on life: without life; ideas such as signal and response and autonomy just don't make sense.
When considering autonomy (e.g. Farnsworth 2017), I noted that at the cellular boundary, cause and effect were converted into signal and response by a combination of the boundary (providing isolation) and transducers (which allow information but very little physical cause through). The only way this can be achieved is through causal isolation and that is what the boundary is for (it keeps the cell's contents in as well, of course). The boundary maintains a difference between physical and chemical conditions: inside different from outside the cell. The specific structure of the boundary allows selective transmission of chemicals and limited forces to pass through. The boundary includes transducers which largely strip efficient causes of their amplitude, but transfer information about their variation through to the cell. Recalling that efficient causes are constrained physical forces, we can see the action of transducers, embedded within the cellular boundary, as constraint of external forces. This constraint is physically realised by the configuration of matter constituting the transducer. The configuration is the particular molecular shape and composition of the transducer.
The translation system of a physically realised code is itself a kind of transducer since it removed physical force from the information that it carries - information resulting from its correlation with the constraints that were placed upon it. The semaphore flags are placed in a configuration by physical forces, but those forces have no action upon the system that responds to the sign they convey.
This is clearly demonstrated in another biological code - cell signaling via transmembrane proteins. The transmembrane proteins act as transducers which relate receptors and their signals (first messenger) to internal information signals (second messenger) of the cell. These second messenger signals transmit through a different, internal, chemistry that is isolated from the extra-cellular environment. Transmembrane proteins can have one of a range of receptors on the outside and a range of second messenger signal molecules on the inside, so again are a translation of information from one kind to another.
Not only is the coded relationship between one set of signs and another arbitrary in a relational code, so are the sets of signs. For example, if we go back to the semaphore, the set of flag positions is an arbitrary choice and so also is the set of letters in the Latin alphabet. The code relates one arbitrary sign uniquely to another. As the difference between RNA and DNA shows (and there are many more examples in synthetic life), the set of four nucleotides is not inevitable - alternatives are available. The same is true of amino acids and cell signaling molecules of course.
References
Barbieri, M. (1998). The organic codes. The basic mechanism of macroevolution. Rivista di Biologia-Biology Forum. 91, 481-514.
Barbieri, M. (2003). The Organic Codes. An introduction to semantic biology. Cambridge University Press.
Barbieri, M. (2018). What is code biology? Biosystems 164:1,10.
Farnsworth, K.D., 2017. Can a robot have free will? Entropy 19, 237 DOI: 10.3390/e19050237.
Hofmeyr, J.H.S. (2007). Systems biology: philosophical foundations.. Elsevier, Amsterdam. The biochemical factory that autonomously fabricates itself: a systems biological view of the living cell. Systems biology: philosophical foundations. pp. pp 217?242.
Hofmeyr, J.H.S. (2017). Handbook of Anticipation: Theoretical and Applied Aspects of the use of Future in Decision Making. Springer. chapter Basic Biological Anticipation. 11, pp. 219?233.
Hofmeyr, J.H.S. (2018). Causation, constructors and codes. BioSystems 164, 121?127.
Kühn, S. and Hofmeyr, J.H.S. (2014). Is the "Histone Code" an organic code? Biosemiotics 7:203-222.
Tomkins, G. M. (1975). The metabolic code. Science 189:760-763.
Wang, A. Nairn, N. A., Marelli, M. Grabstein, K. (2012). Protein engineering with non-natural amino acids. Ch.11 DOI: 10.5772/28719 (available from ResarchGate).