Information Bootstrap: the way programs make themselves
Keith Farnsworth, May 2019 - updated 25-02-2021Software
The raw material for information is a set of differences and its elementary unit is a single binary difference (yes | no, or on | off, or true | false). We are used to this idea, since binary data is the raw material of digital computing. A very important example in biology is the molecular switch: usually a protein, for example a G-protein coupled receptor, which reversibly changes shape upon receiving an appropriate signal (see code biology). Every living cell is full of thousands of molecular switches, just as a digital computer is full of electronic switches.
When someone says ‘software’ we probably think of the scripts (often termed 'code') written by people for computers to follow, be they a word-processor, a calculator, an ‘app’ on the smartphone, or the management system of a car engine or a washing machine. A script is a set of instructions that the computer ‘hardware’ follows or carries out. In all digital computers (your laptop, the engine management system in the car engine, etc.) the actions that can be performed and therefore the only valid instructions that can be followed, are acts of switching electrical connections on or off. That is all. It is a marvel of computer engineering that a sequence of switching connections on and off, and nothing more, can amount to a flight simulator, a video to watch, a diary, or the inner workings of a medical diagnostic scanner. We now know the fundamental reason why this is true. All computation is information processing. By processing we mean taking bits of information and combining them, to make new information. By information, we mean, at the most atomistic level, binary differences: true and false, or on and off. Thus it is that information can always be represented by a set of on and off states: a binary sequence (see Information from Uncertainty). By the same token, information processing can always be reduced to the changing of those on and off states by reversing some of them. This is why digital computers are so powerful and have become so ubiquitous in modern life. The only limitation seems to be the ingenuity of the programmer to reduce a problem to a set of on-off sequences and working out how changing these will achieve the required outcome (the new information).
To be clear, the software for digital computers is itself information: the set of instructions is itself a sequence of on and off. Given this, it is perfectly reasonable to say that a program may itself be the result of information processing: that is a new set of instructions can be the output from following a previously established set of instructions. So the instructions, however coded, can, when followed, result in a set of instructions that themselves are the instructions that when followed are the instructions ... etc. in an infinite loop. This information self-replication idea is captured in Von Neumann's self replicating universal constructor and an example of one in action, built within the coding rules of John Conway's Game of Life can be found here. [Note it is an automaton using the 32-state extended von Neumann rules and was written and published by Golly (golly.sourceforge.net)]. The critical point about the Von Neumann self replicator is that it is both a self-copying program and it also contains the instructions needed to decode (read and interpret) the program, i.e. to run itself.
In other words the self-replicator is a program that computes itself. But looking at the Golly example suggests that this, though very clever, is itself a cheat because it really is quite a complicated and information rich thing. Once in existence, it can replicate itself autonomously, but how did it get there in the first place? The answer of course is that somebody had to program it in. To get over that problem, to truly have an autonomous program, we have no choice other than to turn to the idea of bootstrapping.
Making oneself
When you start up your computer (e.g. smartphone) the first thing it does is 'boot up'. This term dates back to the early years of computing and it was explicitly illustrated it in the computers of the 1970s. I remember one which occupied an equipment rack two metres tall, and along its front was a row of toggle (on/off) switches. These were set to make a binary sequence (e.g. 100111010101) that was a very small amount of information, but it was enough to provide an electronic instruction that combined with a permanently stored small piece of information could start the most basic elements of the operating system in software, which in turn unwrapped more complex and information-rich algorithms and slowly the whole software of the operating system emerged. This was called bootstrapping (a reference to the apparent paradox of lifting oneself up by the bootstraps). It is a form of self-assembly: not of material patterns (as in molecular self assembly), but of pure information. Of course there was some 'cheating' involved because information was being read (incorporated) from storage in tapes or disks. The cheating is good in this case because it is good engineering design. But we are primarily concerned here with how life does this job and we assume there was no engineer to design it.
Extant life does indeed cheat like this via reproduction. Obviously a critical element of reproduction (very much including cell division) is the copying (replication) of information stored in the medium of DNA. But this leads directly to another chicken and egg problem of self-making and the need for an information bootstrap. Put simply: if the instructions to make and 'run' a cell are coded in DNA and that includes the instructions on how to decode (read) the DNA, then given just a DNA program, how is it to be read? It cannot be because until it is read, the way to read it is unknown and without knowing the way to read it, it cannot be read. The way out of this conundrum for all cells today is that they also cheat by storing the decoding (how to read) instructions separately in transfer RNAs and their accompanying aminoacyl-tRNA synthetases (see code biology for details). These relatively small molecules are part of the cystol (the soft jelly of the cytoplasm) and collectively carry the boot-up information enabling DNA to be read and used to make functioning proteins and run the cell.
Very good, but that does not really explain how a program can write itself, which must be what happened in the case of the very first life-form. It had no other life form from which to copy or inherit DNA, RNA or even tRNA molecules. The beginning of life itself had to involve the original writing of a program that not only copies itself, but also carries with it the instructions on how to read, interpret and follow its instructions. The fact that that is at all possible is demonstrated by our living presence. That it has never been observed (not even close) in any system other than life is probably testament to just how hard this computation problem is. Indeed, it may be so difficult that it has only ever been solved by life, but then if any system were to solve it, perhaps we would identify that system as life.
So back to this program that modifies itself, remembering that both the program and its outcome are reducible to a series of on/off states of switches. A very simple, but terribly inefficient, way to write a program is by random selection through trial and error. Writing on the Niche Construction website, Bruce Damer provides an illustration of this approach (Part 2. Programming without a programmer). He imagines a computer with the task of lighting up some LEDs on its front panel. It takes coded messages from a paper tape (in which holes punched represent instructions, using an alphabet of binary bytes (hole=1, no hole =0). Random codes are made with a random tape puncher. The tape is fed into the computer and most of the time the codes are nonsense and end up in the rubbish bin. At random, eventually a code is produced that turns on a light (first little success). That code is (somehow) retained and we continue with random codes, now being added to the first working code. Gradually more randomly generated codes turn out to work, are retained and the correct program to switch on all the LEDs emerges.
OK, what is wrong with this? First there is no word on how correctly working bits of code are retained, rather than binned as all the rest is. Second, the computer reading the code has to be able to do something with it and that something is to switch on and off (according to the instruction) parts of the circuit that lead to the LEDs. This is very easy to do (it is the principle of the very first programmable machines which were Jacquard looms: the things that inspired Charles Babbage to invent a very early computer which he called the Analytic Engine - see this webpage). But the machinery to do it has to be designed and built. The process of designing and building the whole apparatus is an act of embodying information. That information is necessary for its operation, indeed it defines its operation. Crucially none of that information is put together by the apparatus, so it turns out to be a red herring. To be fair, this is exactly the point really. There are no imaginable systems (yet) that can do the job that life did, of autonomously implementing a von Neumann self-replicator. However the idea of selecting from random generation, those things that work - that idea is spot on.
Making new information
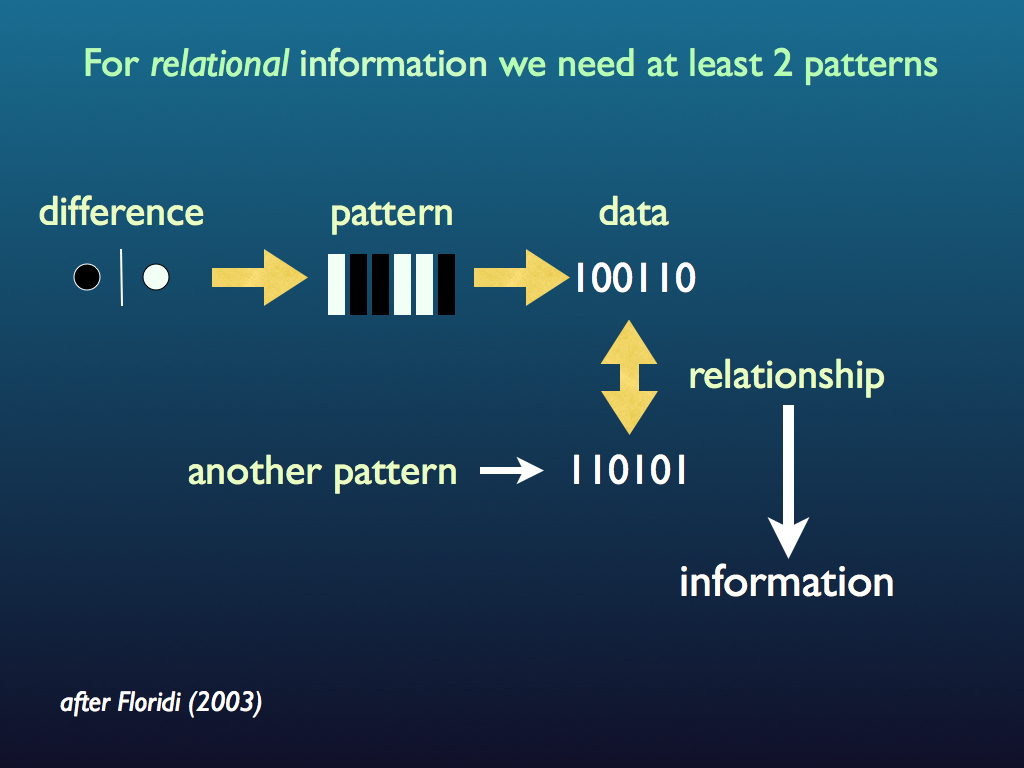
Referring to the diagram at the top of this page again, we see that information is created from the interaction among two (or more) patterns (recall that information is relational). To make new information, two pieces of prior information have to interact. That is why theories about the origin of life tend to need (at some stage) a template for replicating information (see the sequence paradox page). Foremost among these theories is the RNA-first hypothesis and that is because RNA molecules can have both information carrying and functional activity. It is in principle possible for an RNA molecule to work as a little machine that both makes RNA molecules and carries the information telling it how to make them - indeed hypothetically, how to make itself. Such a thing, if it were to exist, would be a self-replicating ribozyme. Biologists who can afford to*, are searching for signs of these; they have not found any such ribozymes yet, but there are a lot of encouraging clues. One speculative possibility is that rather than a single unit, the first self-replicating molecular machine capable of evolution was a set of molecules that collectively made one another. This is close to the idea of autocatalytic sets and it may be an answer to the sequence paradox.
* Very nearly all research grants these days are for applied, more or less engineering work that civil servants have dreamt up, emphatically not basic science.
Supposing we had a very basic, but working molecular von Neumann self-replicator. If that was all, then it would never change, it would continue to proliferate until it had used up all its chemical resources and then that would be the end (of course a population of them might be able to persist if recycling parts could be made sustainable). If it was made from RNA, then its self-replication would actually be horribly error prone and all kinds of ugly mutants would be produced, almost all of them would fail. One common kind of mutation would be the addition of extra bits of RNA at random places. Very rarely, one of these would enable self-replication to either be more efficient, less error prone or enable the self-replicator to do something else that improved its persistence, or replication rate - in other words make it fitter in the biological sense. The introduction of random extra RNA is the introduction of new data, which if it matches the pattern of information already in the molecule (in the special sense that together the old and new are functional), then new information will have been acquired out of randomness and should persist within the larger system (the ribozyme). One thing that would be very useful is way to protect the fragile RNA from free-radical damage. In extant life and most clearly in RNA viruses, this service is provided by proteins, the making of which is informed by the RNA. An even more relevant example just now is the full ribosome which is a bundle of RNA and protein molecules. The very first proto-ribosome was most likely much the same as the translational core containing the peptidyl transferase centre of modern ribosomes (Fox, 2010; Petrov et al., 2014; Petrov et al. 2015). Just maybe, these arose from a ribozyme that could make proteins and wrap itself around them to form a proto-ribosome. Maybe also it could make proteins that formed a shell around itself - a protein capsid, which would then make it like a virus, but better than a virus because it would be independently able to complete its replication cycle (I discuss that sort of thing in Farnsworth 2021).
To be clear, it is not possible to create information out of nothing. The only possible source of new information is the filtering of random data. This filtering is a kind of pattern matching, one in which the combination of existing information (pattern) with a new bit of information is selected (thereby reducing system entropy). The selection criterion may be an increase in some objective function, for example biological fitness, or energy minimisation (e.g. in the case of selecting the location of a new atom as it is incorporated into the pattern of a growing crystal). It is this way that the total amount of information in the universe has increased through time.
Booting up is the act of using a small amount of information to collect more until the total has function. In principle that can be achieved, very slowly, by selecting small amounts of data from randomness, step by step. This is what evolution by natural selection does. In reality, all extant life boots up (every new cell) by 'cheating' through taking ready made information from the parent cell, just as a computer boots up by reading its own operating system off a memory store, starting with a very simple program that implements the reading of it. A new cell uses the set of tRNA, aminoacyl-tRNA synthetases and accompanying chaperones and ribosomes that it takes from the cytoplasm of its parent. It takes the DNA from her too, so it can use that information to make new tRNA etc. How the very first living cell managed to accumulate the necessary information for its own operating system remains a mystery for now.
Responding to the environment
Returning to computer programs, we know that a self-replicating program is not much use unless it can respond to its environment in some way (and engineers are working to develop robots informed by such programs that can literally make themselves, especially for operations in very remote environments, such as on Mars).
For this to work, a stream of information from the environment, such as temperature data from a sensor, or images from a camera could enter the computer running the program in such a way that the modifications of its program depend on the incoming data. The result will be that the computer program running on this computer will not be fixed, but instead will depend on the information it receives from the environment. We tend to think of a program as a rigid set of rules that cannot change: now we see that the rules can change and even that the rules themselves can respond to the external environment. What the computer does is then not what the programmer wrote in code in the first place, it does something that is the result of its adaptation to its own environment. Again, living cells provide a very good example of this adaptive information processing system and this aspect of living information processing is elaborated on the autopoiesis page. Permanently i ncorporating environmentally derived information causes organisms to adapt to their environment and thereby gain fitness - this is the mechanism behind evolutionary adaptive radiation that has given rise to the diversity of life.
Self-rewriting programs can be contrived by computer scientists, but do they exist for real? The answer to this question lies in all of us. Indeed it lies in all living things, because this adaptation of a program is an essential feature of life itself. We still don't really know how it was established in the first instance, but we have many of the essential principles and ingredients in place now.
References
Farnsworth, K. D. (2021) An organisational systems-biology view of viruses explains why they are not alive. Biosystems 200, 104324. Doi: 10.1016/j.biosystems.2020.104324.
Fox, G.E., 2010. Origin and evolution of the ribosome. Cold Spring Harbor Perspect. Biol. 2, a003483.
Petrov, A.S., Bernier, C.R., Hsiao, C., Norris, A.M., Kovacs, N.A., Waterbury, C.C., Stepanov, V.G., Harvey, S.C., Fox, G.E., Wartell, R.M., Hud, N.V., Williams, L.D., 2014. Evolution of the ribosome at atomic resolution. Proceedings of the National Academy of Sciences 111, 10251–10256. doi:10.1073/pnas.1407205111.
Petrov, A.S., Gulen, B., Norris, A.M., Kovacs, N.A., Bernier, C.R., Lanier, K., Fox, G.E., Harvey, S.C., Wartell, R.M., Hud, N.e., 2015. History of the ribo- some and the origin of translation. Proc. Natl. Acad. Sci. U. S. A. , 15396–15401 doi:10.1073/pnas.1509761112.
